Epigenomics of CNS Toxins
Bioinformatics is the study of biological problems through the use of computational technologies. This discipline has been greatly facilitated by the explosion of high-dimensional biological data that has been acquired and placed in the public domain in the last 10-15 years. In our lab, we try to integrate public data into our projects in any way possible, to advance our understanding or to generate new hypotheses. Examples of current efforts underway in this area are 1) the investigation of DNA sequence motif enrichment at risk genes, and 2) integration of expression and epigenetic data with disease-associated transcription factor binding profiles.
The figure below gives an example of mapping enrichment or depletion of DNA methylation quantitative trait loci (meQTLs) in different chromatin states. Our interest was to determine if meQTLs that contained polymorphic CpGs (CpG-SNPs) were more likely to be in regions of active or inactive chromatin. Vertical columns indicate common blood cell types, horizontal columns indicate active or inactive chromatin. Regions enriched with meQTLs are in red, versus depleted in blue. The graphic shows that meQTLs with CpG-SNPs are enriched in heterochromatin (het) and quiescent chromatin (quies) and depleted in most active chromatin. Conversely, meQTLs lacking CpG-SNPs were enriched in transcription start sites (tss).
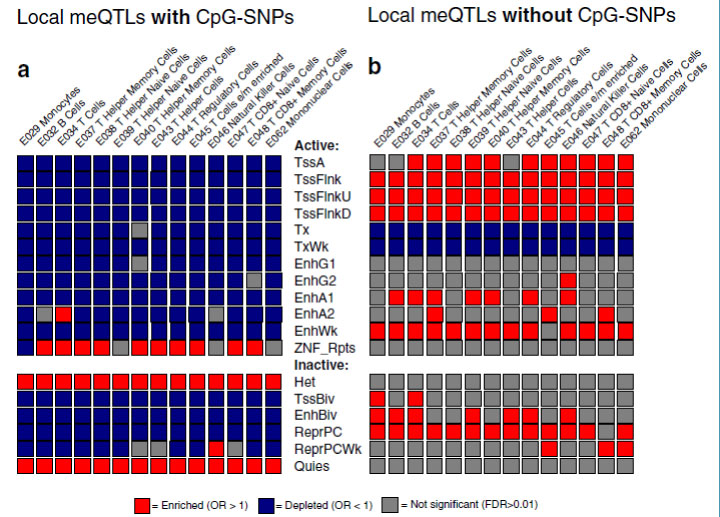
Figure caption: Bioinformatics
Mapping of enrichment patterns for DNA methylation quantitative trait loci in regions of active or inactive chromatin, for 14 different types of blood cell. See McClay et al. Genome Biology 16 (1): 291