Psychiatric Drug Response
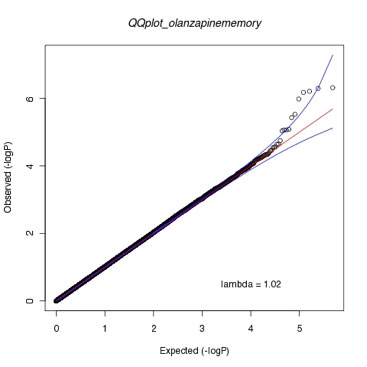
Figure caption:
Quantile-quantile plot showing approximately 492,000 genome-wide p-values for markers mediating response to olanzapine, measured as improvement in neurocognitive functioning (working memory). The QQ plot shows the deviation of markers from the expectation under the null (red line) and 95% confidence intervals (blue lines). See McClay et al. Neuropsychopharmacology 36 (3): 616-626.
No single psychiatric drug works in every patient. For every class of drugs, including antipsychotics, antidepressants, mood stabilizers, etc., there are substantial differences in how individual patients respond and what side effects they experience. Research in recent decades suggests that a substantial portion of these individual differences in response are due to genetic differences between patients. This has spurred the hunt for genes that influence drug response, which has led to the emergent discipline of pharmacogenetics/genomics (PGx).
Until recently PGx research focused on candidate genes, where genes were selected based on their coding for drug metabolizing enzymes (pharmacokinetics) or proteins involved in the drug’s mode of action (pharmacodynamics). In psychiatry, this approach has had limited success. Current FDA-approved biomarkers for psychiatric drugs include just two genes that encode drug metabolizing enzymes and no genes at all involved in pharmacodynamic processes. However, recent technological advances now allow us to screen the entire genome for genes influencing drug response. The PI was involved in genome-wide association studies (GWAS) of antipsychotic drug response in the Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) sample, the largest clinical trial of these drugs funded by the National Institutes of Health. These GWAS studies found genetic markers that differentiated individuals on their response to antipsychotics, based on reduction in schizophrenia symptoms or improvement in neurocognitive functioning. Several of these markers were in genes implicated in other psychiatric PGx studies or associated with risk for psychiatric disorders. We have prioritized some of these genes for follow-up and functional characterization. One of these genes is ANKS1B, which encodes an activity-dependent effector protein of the post-synaptic density. We recently completed the initial characterization of a mouse knockout for this gene and are planning additional biological studies of drug response in this model.
"Omics" studies of psychiatric drug response
We are interested in understanding the biological mechanisms of psychiatric drug response and side effects. To this end, we use functional “omics” technologies such as proteomics and metabolomics. In much the same way as genomics attempts to assay the entire genome, metabolomics and proteomics attempt to assay as many small molecule metabolites or proteins that exist in a sample as possible. In one recent metabolomics study, we examined the effects of long term haloperidol administration in the brain. Haloperidol is a first generation antipsychotic with a high risk for extrapyramidal side effects (EPS), which involve involuntary movements and loss of motor control. In our study, we identified disruptions to several metabolites that pointed to haloperidol-induced demyelination. In the brain, myelin is the fatty insulating sheath that surrounds axon fibers. Our study suggests that long-term haloperidol administration leads to loss of this insulator, thereby interfering with neural signaling. Our next step in this project is to identify if sphingolipid species in the blood can be used as biomarkers to predict EPS prior to symptoms manifesting.
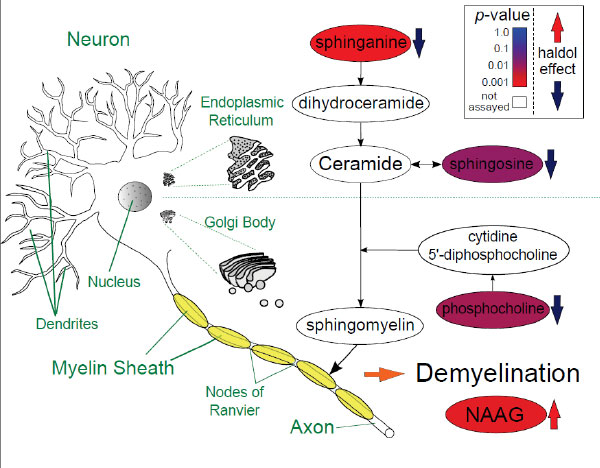
Figure caption: Psychiatric drug response
Schematic showing changes to metabolites in the central nervous system, color coded by their p-values, in a metabolomics study of long-term haloperidol administration. See McClay et al. Journal of Neuroimmune Pharmacology 10 (3): 425-434.